Projects
Determination of sequence-based effects of the 2A peptide on ribosomal skipping efficiency
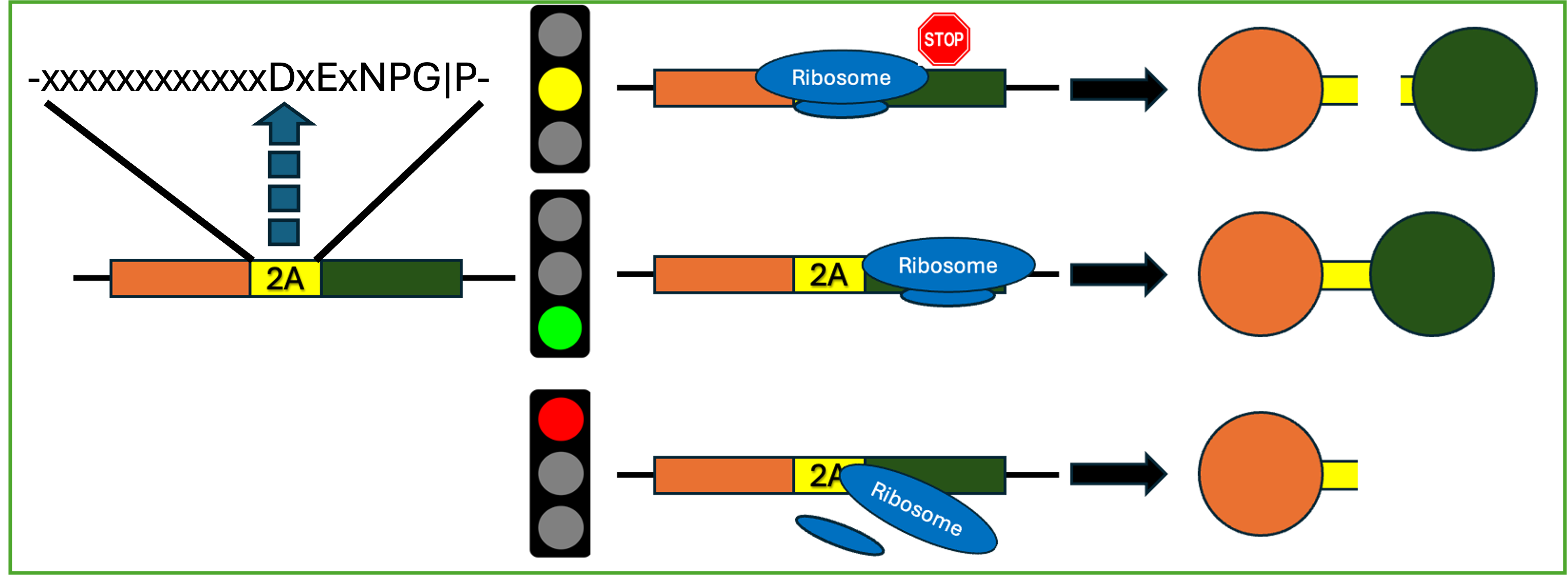
The aphthovirus subfamily of picornaviridae contain a protein sequence called the 2A peptide that is considered to mediate a ribosomal ‘skipping’ mechanism. This mechanism takes 3 forms: skipping, where failure to form the G-P peptide bond occurs and two proteins are produced from a single translation event; readthrough, where the G-P peptide bond is formed and a fusion product is produced; and falloff, where failure to form the G-P peptide bond occurs, and translation terminates, producing only one protein product. These events are considered to occur at some specific ratio based on the peptide sequence. At present, it is known that a C-terminal DxExNPGP is conserved among sequences that have been observed to mediate skipping. However, the 20 residues culminating in the C-terminal NPGP are observed to be necessary for skipping to occur, and are broadly unconserved among sequences observed to mediate skipping. In order to understand how upstream sequence mediates ribosomal skipping, we are utilizing site saturation mutagenesis approaches to produce the largest libraries of 2A and 2A-like sequences to date.
Survey Potential Ribosomal skipping in diverse Eukaryotic 2A-like sequences
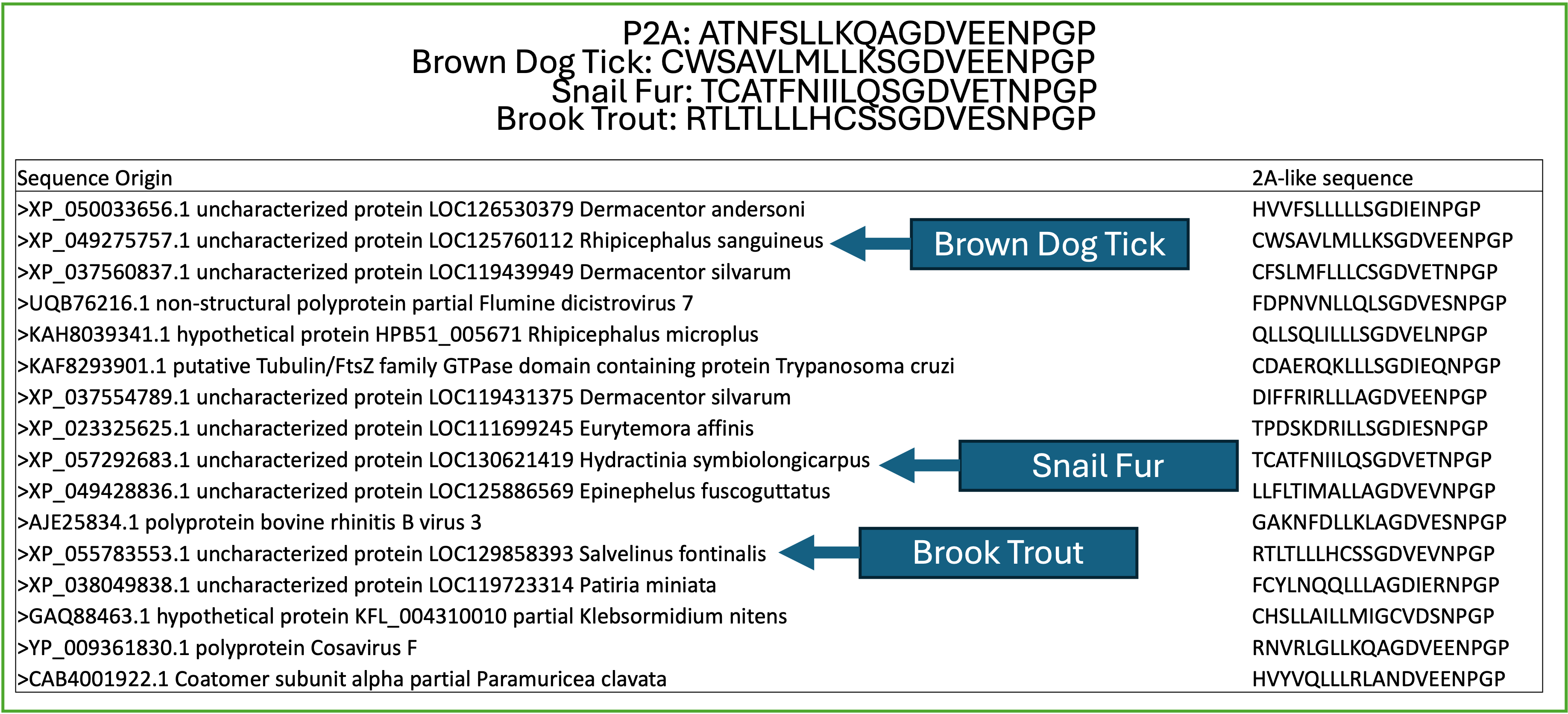
When performing precursory BLAST searches utilizing the most studied 2A peptides, Eukaryotic sequences of unknown function frequently arise. Given their similarity to aphthovirus picornaviridae 2A peptides, we seek to determine if a previously undescribed “ribosomal skipping” function motif exists in Eukarya, which we are determining through high throughput analyses.