Additional Data From The Kozak Paper
Recently, our lab published a preprint paper on kozak sequences linked here. My contribution to this paper involved finding and defining the clinvar variants in kozak sequences. Outside of this, I had also done some other work to consider mutations in cancer (which did not make it into the paper), and kozak sequence frequencies in other species using an adjacent methodology (also did not make it into the paper). I wanted to share some of that data here.
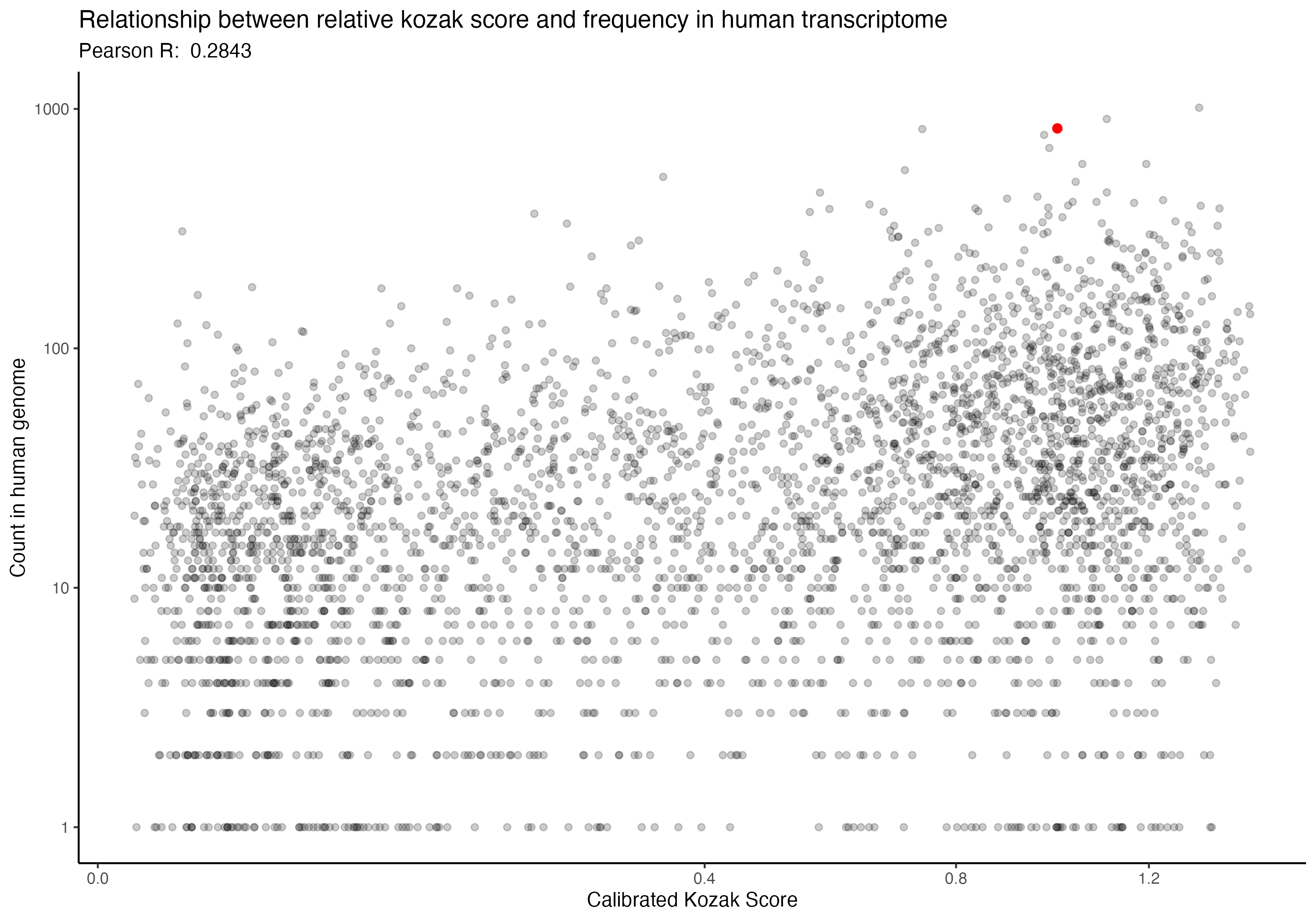
For the following graphs, please note that the red dot identifies the ‘consensus kozak’ of GCCACCATG.
First, because I think it is interesting, this is the correlation of our calibrated scores and kozak sequence frequency in the human genome.
Next, a series of graphs, documenting the correlation of kozak frequency between humans and a number of other species. Plots are ordered based on my filename order.
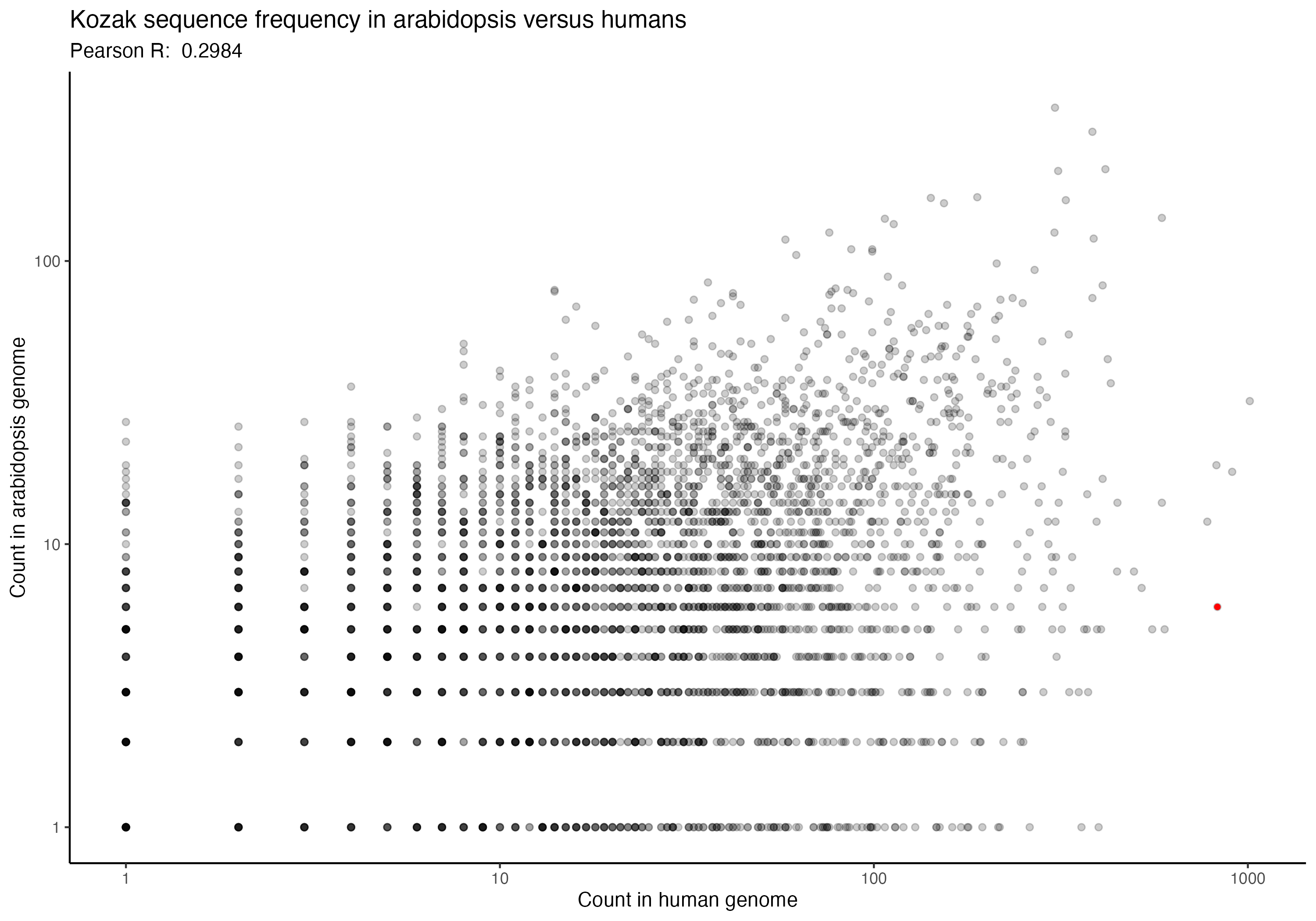
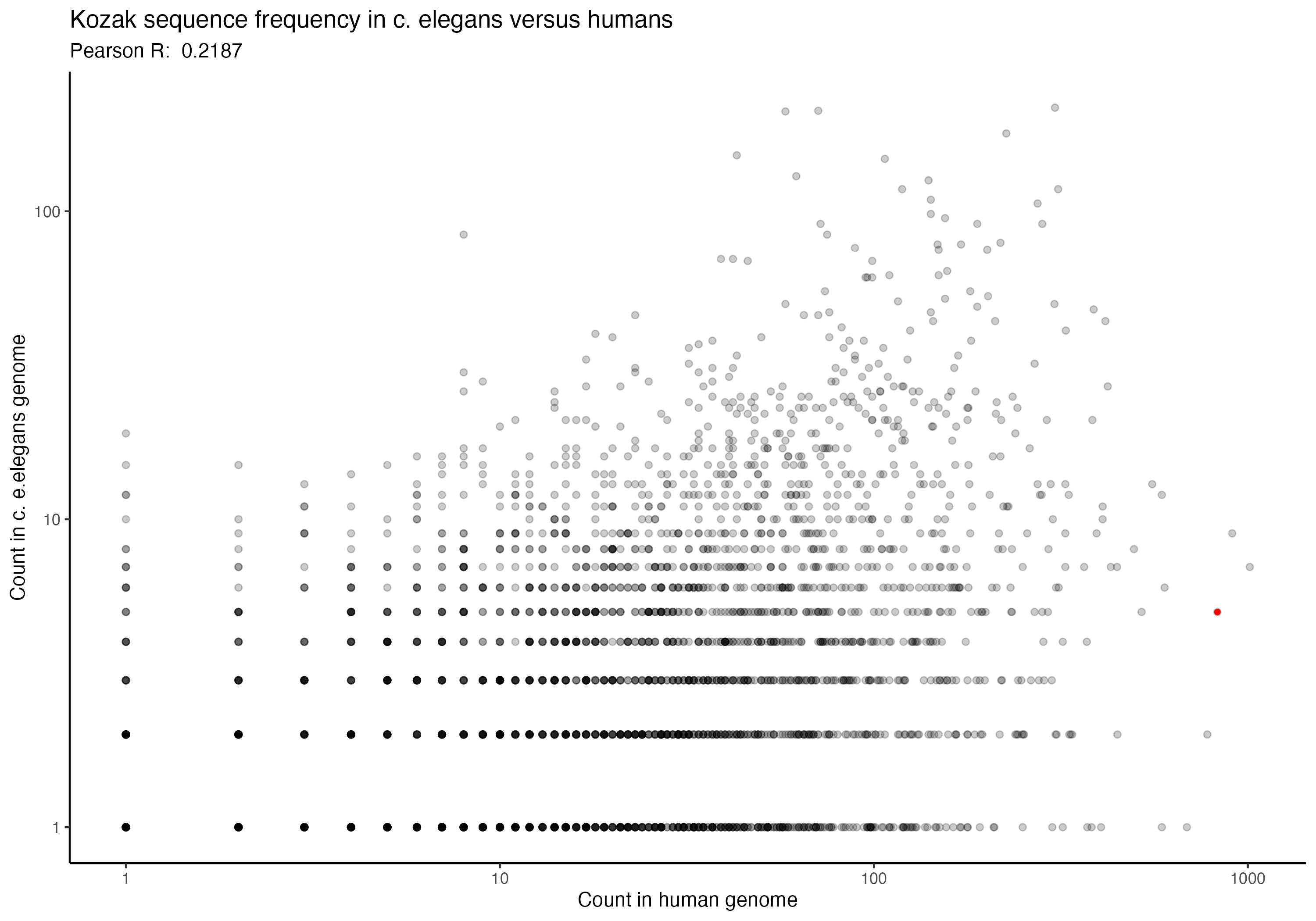
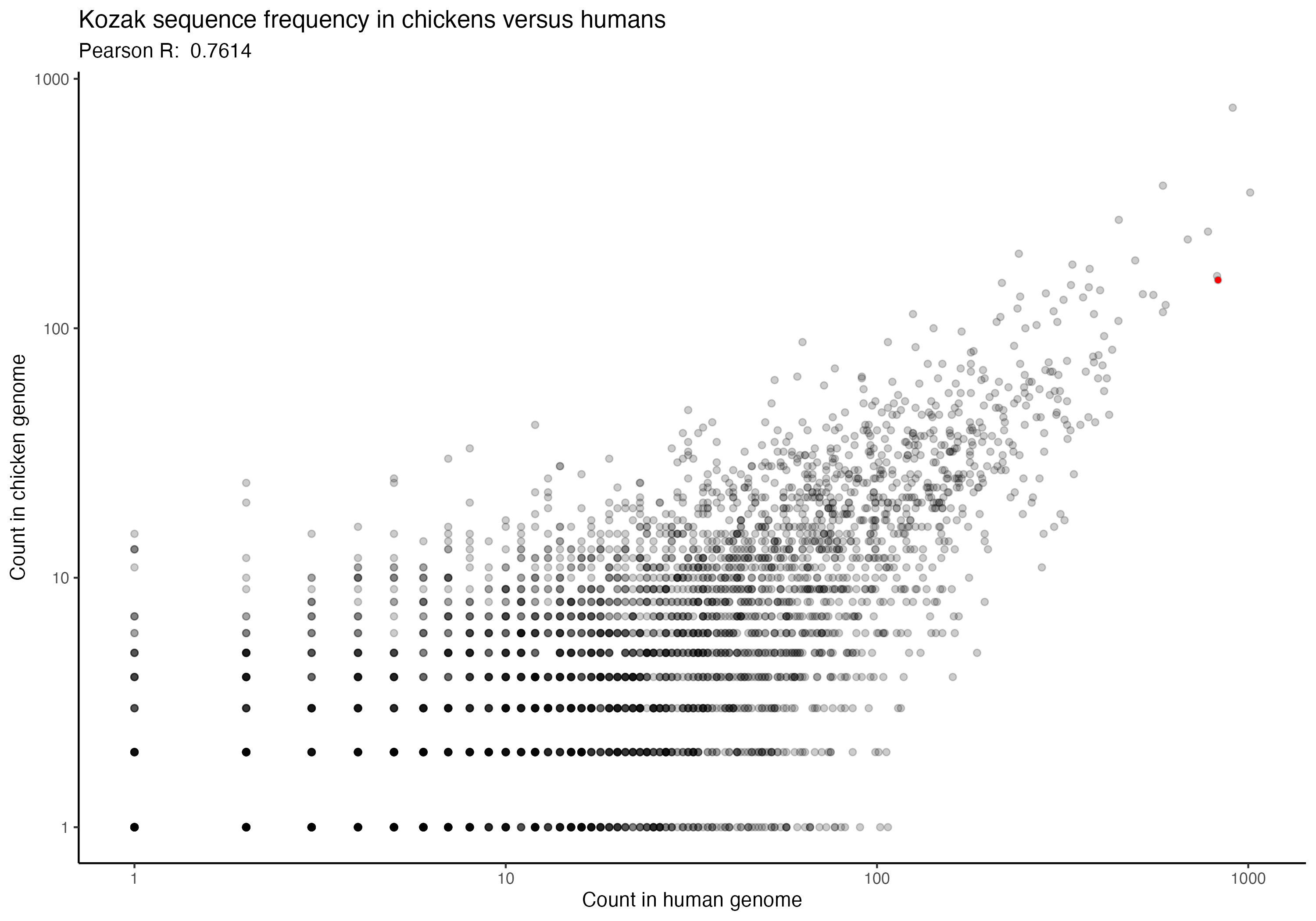
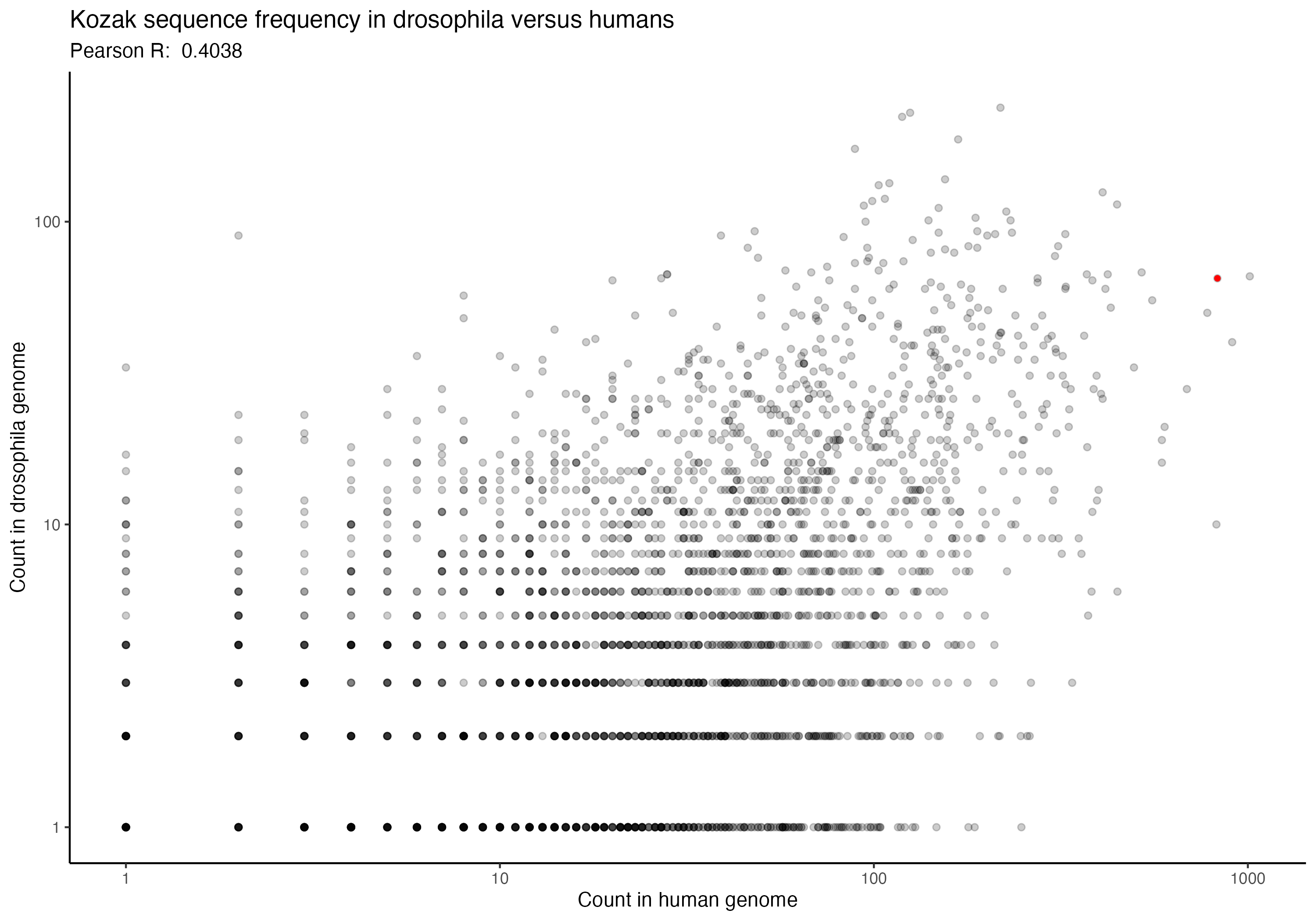
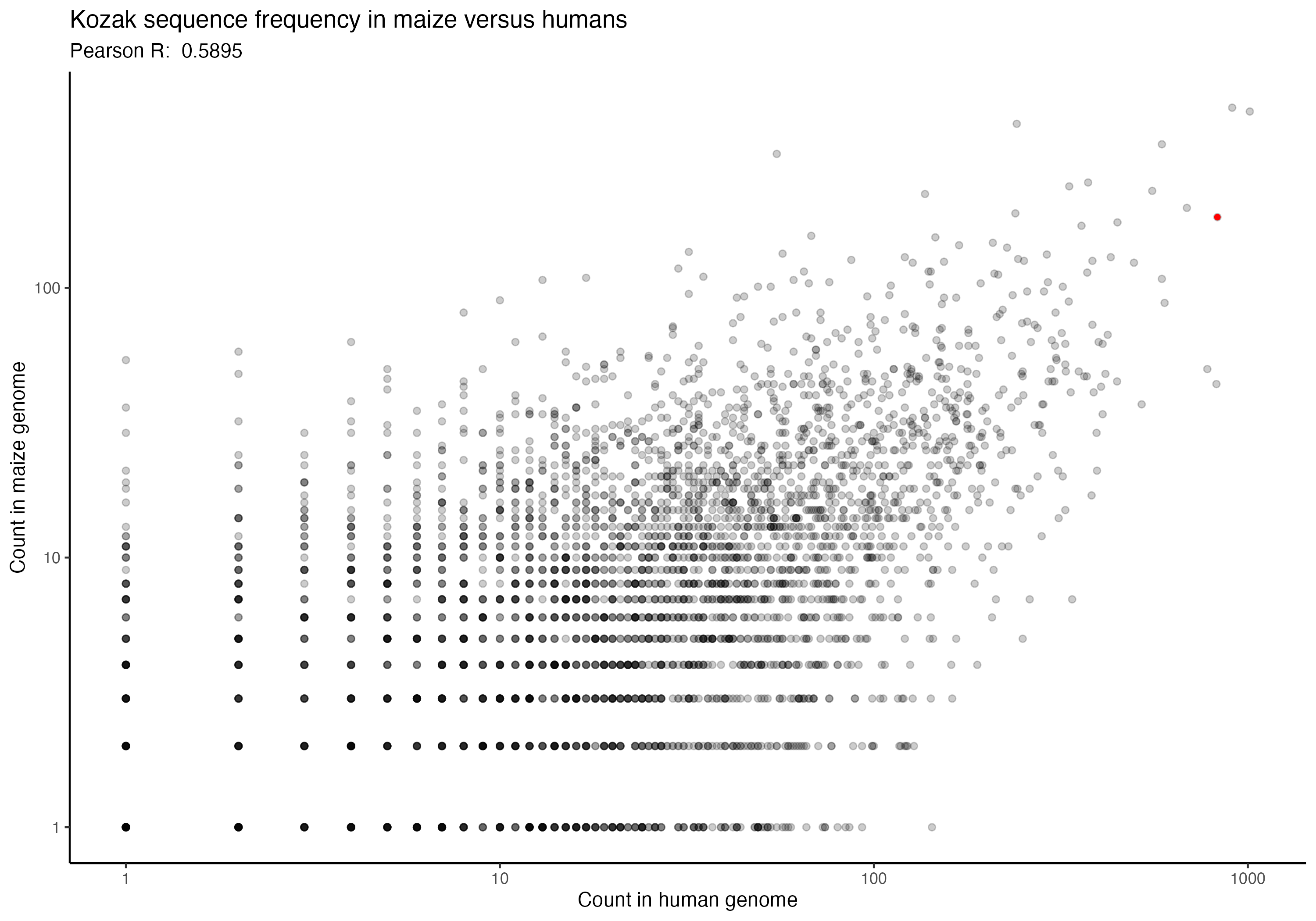
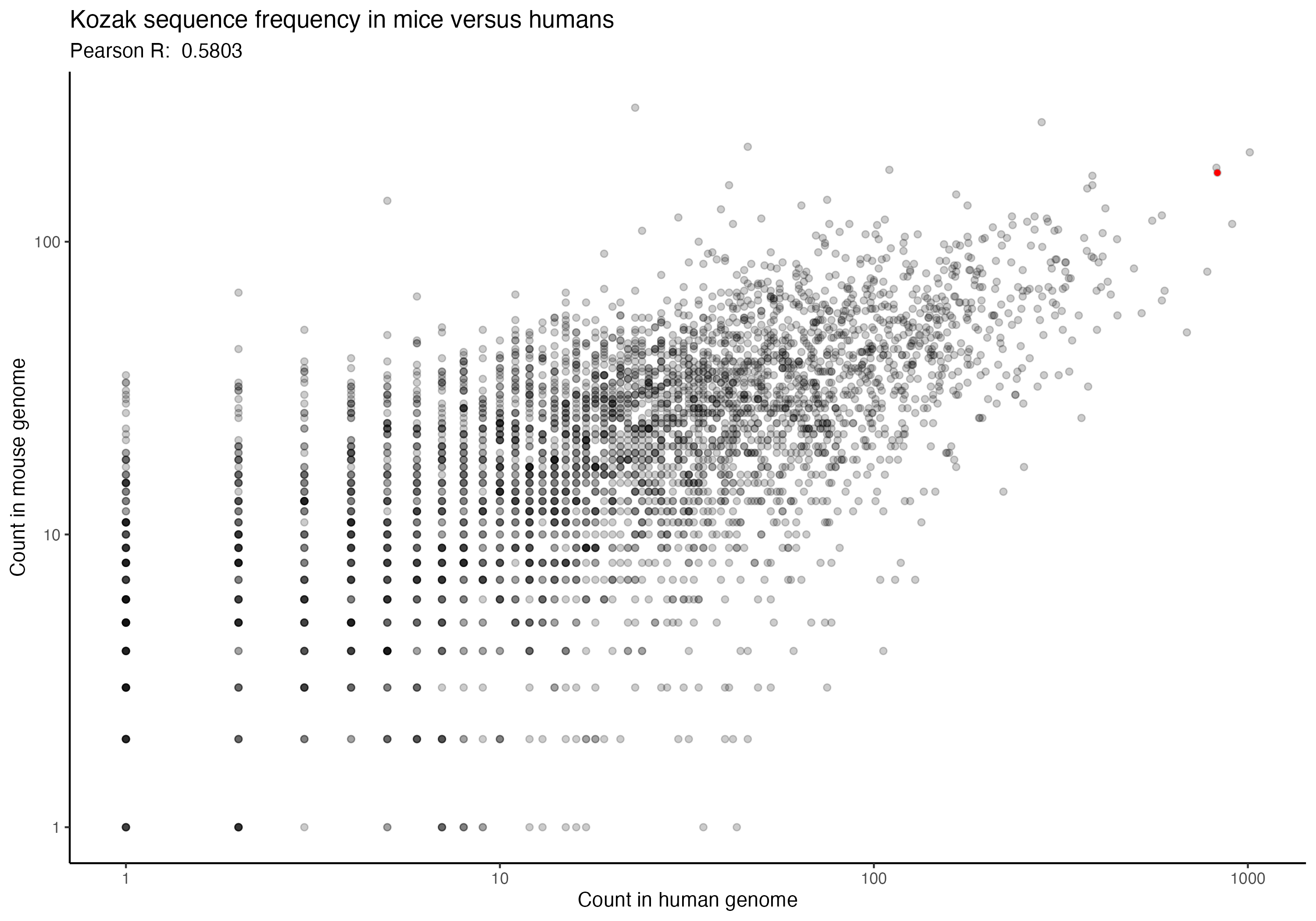
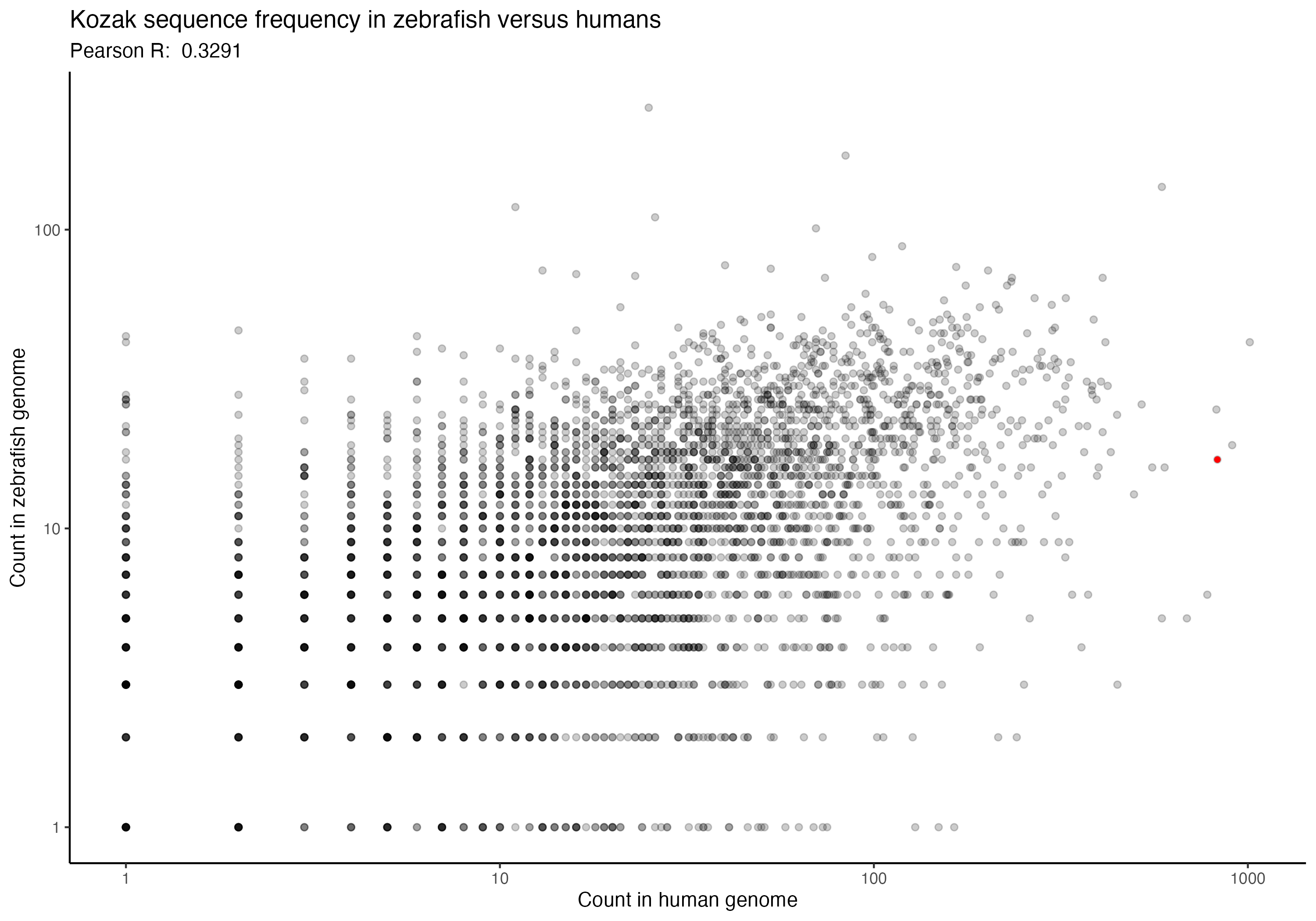
(underlying data is available upon request)
In going back to these (data is ~2 years old now), I was hoping that I might spot some interesting trend that could perhaps allow some extrapolation forward, but I don’t see any particular trend that stands out immediately. Chances are, the kozak order and effect for human cells (from the paper) is likely to be generally applicable across species, but would be good for someone to confirm someday. If anything, I imagine that setting up an arabidopsis or zebrafish landing pad system wouldn’t be terribly difficult, although I also imagine that they aren’t likely to be great candidates for many practical applications; however, a mouse or maize landing pad being built to check that the kozak sequences show similar effects in other species could have downstream impacts of not only being good for confirming the kozak data more broadly, but also in potentially allowing further agricultural genetics and biomedical work to proceed. Anyway, if anyone would be interested in that, let me know. I’m rambling, so time to move along.
Ciao